Community Articles
- Cloudera Community
- Support
- Community Articles
- Deep Learning in Medicine: Classifying Melanoma
- Subscribe to RSS Feed
- Mark as New
- Mark as Read
- Bookmark
- Subscribe
- Printer Friendly Page
- Report Inappropriate Content
- Subscribe to RSS Feed
- Mark as New
- Mark as Read
- Bookmark
- Subscribe
- Printer Friendly Page
- Report Inappropriate Content
Created on 10-22-2018 01:20 PM - edited 09-16-2022 01:44 AM
Deep learning offers the opportunity to improve clinical
outcomes in medicine, enabling healthcare providers to serve growing patient
populations, and providing care givers with the
tools they need to focus on patients with critical conditions.
The applications for the use of deep learning in medicine extend across all areas where visual diagnosis is used in patient care. This includes radiology (arteriography, mammography, radiomics), dermatology, and oncology, as well as clinical and biotech R&D.
In this series, we will explore these applications, including the staging of data, training of deep learning models, evaluating models, the use of distributed compute, and the operationalization of our models.
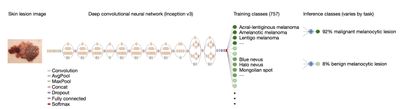
In this article, we will take a look using deep learning to classify Melanoma. Specifically, we will use images of skin, to classify whether the patient associated with the image, has a malignant tumor. The diagram below shows a classification model based on Stanford Medicines classification of skin disease using the Inception v3 Convolutional Neural Network architecture (reprinted from https://research.googleblog.com/2016/03/train-your-own-image-classifier-with.html, and https://cs.stanford.edu/people/esteva/nature/ )
We will explore the use of a pre-trained Convolutional Neural Network (abbreviated ConvNet, or CNN) based on the VGG16 CNN architecture, and trained on the ImageNet data set, and we will retrain that CNN to classify melanoma, using transfer learning.
Numerous CNN architectures are available today, specifically designed for image classification. These include, ResNet, Inception, and Xception, as well as VGG16. While training these CNN’s from scratch requires significant computational resources, and significant datasets, pre-trained weights are available for each as a part of open source libraries, allowing us to build performant classifiers, with significantly reduced input data sets. This means that as clinicians, health care providers, and biotechnology researchers, it is possible to achieve results even with smaller data sets.
In our example we will explore building a classifier with a limited data set of several thousand images.
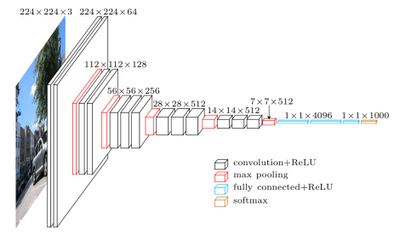
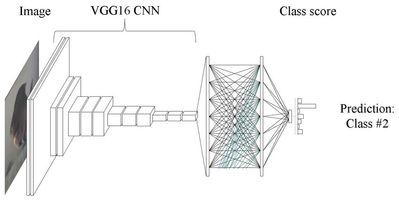
The VGG16 architecture we will be using is shown in the diagram below.
In a pretrained VGG16 model, the convolutional layers towards the visible layer of the network, have already learned numerous tasks necessary for image recognition, such as edge detection, shape detection, spatial relationships, etc.
The top 5 layers of the network, have learned much higher levels of abstraction, and at the top of the network, are performing the classification of our images.
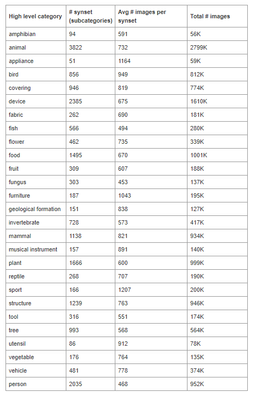
Pretrained VGG16 networks available in open source deep learning libraries such as Keras are typically pretrained on the ImageNet dataset. That dataset comprises of 14 million images open sourced images, none of which include skin samples. The broad classifications for these images are included below.
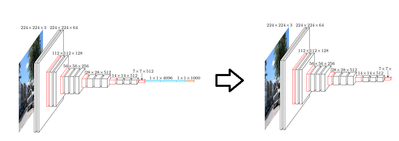
In order for us to build a model suited for a specific task such as classifying melanoma, we will need to retrain VGG16, to perform this more specific classification.
The first thing we will do is remove the top 5 layers of the pre-trained VGG16 model, and keep only the convolutional layers. Remember, the top layers have been trained to recognize features of classes such as animals, plants, people, and objects. These will not be needed in classifying melanoma.
Next we will want to replace the top layers we removed with layers that are more adapted to classify melanoma. To do this we will first feed forward our skin images through our “truncated” VGG16 network – i.e. the convolutional layers. We will then use the output (features) to train a fully connected classifier.
After this we will reconnect the fully connected classifier onto our truncated VGG16 network, and we will then retrain the whole model. Once this is done, we will have a CNN trained to classify melanoma. For details on how to implement this, follow the link here: https://community.hortonworks.com/content/kbentry/226464/part-2-implementing-with-tensorflow-and-ker....
Created on 10-23-2018 07:54 PM
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Report Inappropriate Content
Very cool! Great diagrams too 🙂