Support Questions
- Cloudera Community
- Support
- Support Questions
- Load balancing while the fetching of file from a ...
- Subscribe to RSS Feed
- Mark Question as New
- Mark Question as Read
- Float this Question for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Subscribe to RSS Feed
- Mark Question as New
- Mark Question as Read
- Float this Question for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Load balancing while the fetching of file from a standalone NIFI instance doing the processing it in a NIFI cluster.
- Labels:
-
Apache NiFi
Created 08-24-2016 01:23 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
We have a NIFI setup where in we have a NIFI cluster installed in a hadoop cluster and a standalone NIFI instance running on another server.The input file will be generated in the file system of the server of the standalone instance.We are fetching the file using ListFile/FetchFile processor in the standalone instance.Then in the main cluster we are connecting to the standalone instance using RPG group and then send the output to the NIFI cluster(RPG) using site-site.As per my understanding the part of the processing done inside the cluster will be distributed.Is this understanding correct?I also would like to know if there is a way to distribute the fetching of the source file that we are doing in the standalone NIFI instance?
The flow we are using
In the standalone instance
ListFile->FetchFile->Outputport
In the NCM of the cluster
RPG(Standalone NIFI Instance)-->RPG(NIFI cluster)
Inputport->Routetext->PutHDFS(This is the processing done in the main cluster)
Let me know if you need any other information.Any inputs will be appretiated.
Regards,
Indranil Roy
Created on 09-06-2016 04:45 PM - edited 08-19-2019 03:43 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
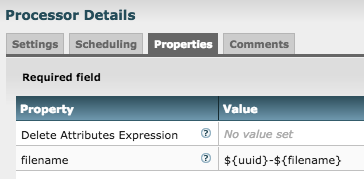
The output from the SplitText and RouteText processors is a bunch of FlowFiles all with the same filename (filename of the original FlowFile they were derived from.) NiFi differentiates these FlowFiles by assigning each a Unique Identifier (UUID). The problem you have is then writing to HDFS only the first FlowFile written with a particular filename is successful. All others result in the error you are seeing. The MergeContent processor you added reduces the impact but does not solve your problem. Remember that nodes do not talk to one another or share files with one another. So each MergeContent is working on its own set of files all derived from the same original source file and each node is producing its own merged file with the same filename. The first node to successfully write its file HDFS wins and the other nodes throw the error you are seeing. What is typically done here is to add an UpdateAttribute processor after each of your MergeContent processor to force a unique name on each of the FlowFiles before writing to HDFS.
The uuid that NiFi assigns to each of these FlowFiles is often prepended or appended to the filename to solve this problem:
If you do not want to merge the FlowFiles, you can simply just add the UpdateAttribute processor in its place. YOu will just end up with a larger number of files written to HDFS.
Thanks,
Matt
Created 08-24-2016 01:49 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
Whenever you do site-to-site, it will automatically do load-balancing for you... So if you have your standalone doing ListFile -> FetchFile -> OutputPort then on your cluster, all you need to do is have a RPG pointing back to the Output Port on the standalone instance, there will be an instance of this RPG on each node of your cluster and each one will pull from the Output port. No need to do another internal RPG.
See the site-to-site section here:
https://community.hortonworks.com/articles/16120/how-do-i-distribute-data-across-a-nifi-cluster.html
You can't really distribute the fetching of the source file if it is coming from the local filesystem. The only thing that can fetch that file is something with access to the local filesystem, which is your standalone NiFI. If it was a shared filesystem then you could.
Created 08-25-2016 06:03 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
Thanks @Bryan Bende for your input
As per your suggestion I have included two different flows as given below:
Flow1
=====
In the standalone instance
ListFile->FetchFile->Outputport
In the NCM of the cluster
RPG(Standalone NIFI Instance)-->Routetext->PutHDFS(This is the processing done in the main cluster)
Flow2
=====
In the standalone instance
ListFile->FetchFile->Input port of RPG(NIFI Cluster URL of the NCM)
In the NCM of the cluster
Input port->RouteText->OutputPort
Which according to you is the correct flow.I can understand that the fetching of the source file cannot be distributed if the file is not shared but which of the flows will be apt to distribute the part of the processing done inside the cluster?
Regards,
Indranil Roy
Created 08-25-2016 12:37 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
Both of those flows should have the data distributed in the cluster...
Flow #1 is a pull model where all the nodes in the cluster will pull data from the standalone.
Flow #2 is a push model where the standalone will push the data to all the nodes in the cluster.
Either approach is correct, but I tend to lean towards the push model (#2) since it lets the source of the data (the standalone instance) decide where to send the data.
Created on 08-24-2016 02:14 PM - edited 08-19-2019 03:44 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
What you describe is a very common dataflow design.
I have a couple question for clarity.
RPG (Remote Process Group) do not send to other RPGs. RPG send and pull data from input and output ports located on other NiFi instances. I suspect your standalone instance has the RPG and it is sending FlowFiles to input port(s) on the destination NiFi cluster. In this particular case the load-balancing of data is being handled by the RPG. For network efficiency data is distributed in batches, so you may not see with light dataflows and exact same number of FlowFiles going to each Node. Also the Load-balancing has logic built in to it so that Node in the target cluster who have less work load get more FlowFiles. Although the URL provided to the RPG is the URL for the target BNiFi cluster's NCM, the FlowFiles are not sent to the NCM, but rather sent directly to the connected nodes in the target cluster.
Every Node in a NiFi cluster operates independently of one another working only on the FlowFiles it possesses. Nodes do not communicate with one another. They simply report their health and status back to the NCM. It is information from those health and status heartbeats that is sent back to the source RPG and used by that RPG to do the smart data delivery.
In order to distribute the fetching of the source data, the source directory would need to be reachable by all nodes in the target NiFi cluster. In the case of ListFile/FetchFile, the directory would need to mounted identically to all systems.
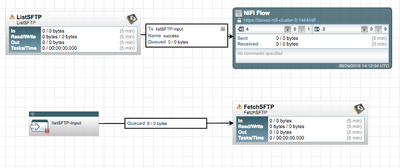
Another option would be to switch to a listSFTP/FetchSFTP setup. In this setup you would not even need your standalone NiFi install. You could simply add a listSFTP processor to your cluster (configured to run "on primary node"). Then take the success from that listing and feed it to an RPG that points back at the clusters NCM URL. An input port would be used to receive the now load-balanced FlowFiles. Feed the success from that input port to the FetchSFTP processor and now you have all nodes in your cluster retrieving the actual content.
So as you can see from the above the listSFTP would only run on one node (Primary Node) producing no content FlowFiles. The RPG would smartly distribute those FlowFile across all connected nodes where the FetchSFTP on each Node would retrieve the actual content.
The same flow above could be done with listFile and FetchFile as well, just mount the same source directory to every node and follow the same model.
Matt
Created 08-25-2016 12:39 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
Hi @mclark Thanks for the alternate approach that you suggested.It could be helpful in my case.
Say in the scenario mentioned above we have a single input file of size in the order of TB's.If we use a ListSFTP/FetchSFTP processor in the way you mentioned to distribute the fetching of data:
- Do we need to establish a SFTP channel between every slave node of the cluster and the remote server that houses the source file for this approach to work?
- Is it a good idea to use SFTP to fetch the file considering the size of the file will be in TB's?
- What are the parameters on which the the performance of the fetch using ListSFTP/FetchSFTP will depend?
Created 08-25-2016 02:00 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
The massive size of your file, ListSFTP/FetchSFTP may not be the best approach. Let me ask a few questions:
1. Are you picking up numerous files of this multi-TB size or are we talking about a single file?
2. Are you trying to send the same TB file to every Node in your cluster or is each node going to receive a completely different file?
3. Is the directory where these files are originally consumed from a local disk or a network mounted disk?
Created 08-25-2016 04:02 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
@mclark
1)We are talking about a single file in TB.
2)There is a single file and the processing should be distributed.
3)The file are in the local directory.
So is it a good idea?
Created 08-25-2016 08:41 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
NiFi does not distribute processing of a single file across multiple Nodes in a NiFi cluster. Each Node works on its own set of files. The Nodes themselves are not even aware other nodes exist. They work on what files they have and report their health and status back to the NiFI Cluster Manager (NCM).
1. What format is this file in?
2. What kind of processing are you trying to do against this files content?
3. Can the file be split in to numerous smaller files (Depending on the file content, NiFi may be able to do the splitting)?
As an example:
A common dataflow involves processing very large log files. The large log file is processed by the SplitText processor to produce many smaller files. These smaller files are then distributed across a cluster of NiFi nodes where the remainder of the processing is performed.
There are a variety of pre-existing "split" type processors.
Thanks,
Matt
Created 08-24-2016 03:34 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
To your questions:
- "Will the processing be distributed" - Yes, the incoming flow of data will be evenly distributed to all available NiFi instances in the NiFi cluster. NCM will act as load balancer
- "Distribute the fetching of the source file" - could you elaborate on what you mean by this? In your existing example the Standalone instance is the only one which has access to its local filesystem. How would you prefer to distribute this?