Support Questions
- Cloudera Community
- Support
- Support Questions
- Re: Running a Python script from NiFi
- Subscribe to RSS Feed
- Mark Question as New
- Mark Question as Read
- Float this Question for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Subscribe to RSS Feed
- Mark Question as New
- Mark Question as Read
- Float this Question for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Running a Python script from NiFi
- Labels:
-
Apache NiFi
Created on 10-08-2018 09:38 AM - edited 08-17-2019 09:20 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
I would like your advice in running a Python script via NiFi. A have a script that will send a data access report (based on Apache Ranger data) to data owners every month. As it is now it needs a bunch of arguments and it will ask for a password with getpass. My fellow data engineers asked me to run it from NiFi, so we have scheduled things together in one tool.
I've experimented with NiFi a bit, but I can't seem to get the Python script to even run in a sandbox environment (HDF 3.1.1) in a ExecuteProcess processor.
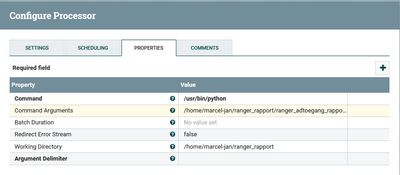
I placed the Python script in /home/marcel-jan/ranger_rapport and even did a chmod 777 on the directory and script, but NiFi says:
'Working Directory' validated against '/home/marcel-jan/ranger_rapport' is invalid because Directory doesn't exist and could not be created.
I just don't get what's going wrong there.
I have a couple of questions:
- Am I using the right processor for this?
- Is it possible to pass a securely stored password on from NiFi to a Python script? If not, what other steps do you take to keep things like this secure?
Created 10-09-2018 08:15 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
@Marcel-Jan Krijgsman
I don't have the same issue, cloning the same steps you've shown on your screenshots in a sandbox HDF. Are you sure you created the directory with the right name and in the right place? Ex when you connect to http://localhost:4200/ with root / hadoop , and do 'll /home/marcel-jan/ranger_rapport' it all looks OK?
Created 10-08-2018 03:07 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
I think I'm getting why NiFi can't find the Python script. Could it be because the node name according to Ambari is sandbox-hdf.hortonworks.com and when I type hostname on the prompt I get sandbox-host.hortonworks.com?
Created 10-08-2018 03:16 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
Hey Marcel:
Something that is helpful sometimes, when trying to troubleshoot why nifi cannot run a process, is dropping into the console of the node that runs the process as the nifi user, and run it from there, to make sure the Nifi User can reach the specific script.
In your specific scenario, it looks like nifi has a problem with the working directory.
Any reason you are not using executestreamcommand for this?
Thanks!
Regards
Created 10-09-2018 08:00 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
Am I right to assume that with dropping into the console you mean starting a Putty session to look on the server? I've done that, but there isn't a nifi user I can su to. I had expected there to be one.
I've also tried a GetFile processor to simply pick up the .py script. Same message: the directory doesn't exist.
I get the same "Working directory doesn't exist" problems with the ExecuteStreamCommand processor.
Created 10-09-2018 08:01 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
It's almost like I'm on a different host with my Putty session. But I honestly have only one sandbox running.
Created 10-08-2018 03:38 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
Hi, Did you try the ExecuteScript processor? You can copy-paste the entire Python Script in the script body property, in case you run into directory access issues.
Created 10-09-2018 07:25 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
I've tried this. I rewrote the Python code so that it won't need arguments and pasted it in the ExecuteScript property. And then it says "cannot use module requests". So I looked into that, and it turns out that you can't install libraries you're missing (https://community.hortonworks.com/questions/53645/cannot-use-numpy-or-scipy-in-python-in-nifi-execut...)
So I think that's off the table.
Created 10-09-2018 08:15 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
@Marcel-Jan Krijgsman
I don't have the same issue, cloning the same steps you've shown on your screenshots in a sandbox HDF. Are you sure you created the directory with the right name and in the right place? Ex when you connect to http://localhost:4200/ with root / hadoop , and do 'll /home/marcel-jan/ranger_rapport' it all looks OK?
Created 10-09-2018 09:36 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
So when you connect to http://localhost:4200/ you get a different hostname than I get when going to sandbox-host.hortonworks.com with SSH and my script wasn't there. I found out that NiFi runs inside a Docker container in the virtual machine. In that sense it is indeed a different machine.
So I placed my script in a directory via http://localhost:4200/ and now NiFi is able to find it.
Created 10-09-2018 10:07 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
@Marcel-Jan Krijgsman
Ah good, glad it works correctly now. When you connect with putty, using port 2222 should bring you to the docker container directly. Otherwise you may be able to do a docker attach to the running docker image.
Please mark the answer as accepted if you can, so others looking for this can find the solution more easily 🙂