Community Articles
- Cloudera Community
- Support
- Community Articles
- Apache Ambari Workflow Manager View for Apache Ooz...
- Subscribe to RSS Feed
- Mark as New
- Mark as Read
- Bookmark
- Subscribe
- Printer Friendly Page
- Report Inappropriate Content
- Subscribe to RSS Feed
- Mark as New
- Mark as Read
- Bookmark
- Subscribe
- Printer Friendly Page
- Report Inappropriate Content
Created on 02-12-2017 03:45 PM - edited 08-17-2019 02:35 PM
In this tutorial, we're going to run a Pig script against Hive tables via HCatalog integration. You can find more information in the following HDP document http://docs.hortonworks.com/HDPDocuments/HDP2/HDP-2.5.3/bk_data-movement-and-integration/content/ch_...
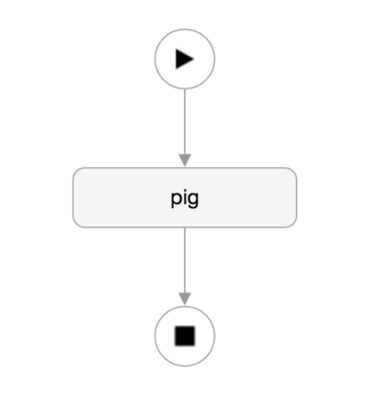
First thing you need to do in WFM is create a new Pig action.
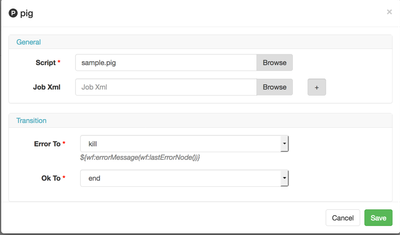
Now you can start editing the properties of the action. Since we're going to run a Pig script, let's add that property into the wf.
This is just saying that we're going to execute a script, you still need to add the <file> attribute to the wf.
This expects a file in the workflow directory, we will need to upload a pig script later.
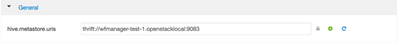
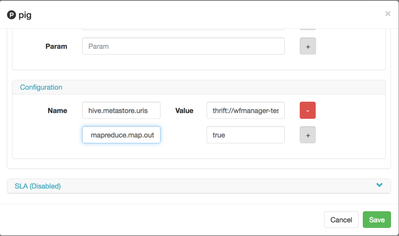
Next, since we're going to run Pig against Hive, we need to provide thrift metastore information for it or include hive-site.xml file into the wf directory, since that usually changes, it's probably best to add the property as part of wf. You can find the property in the Ambari > Hive > Configs and search for hive.metastore.uris.
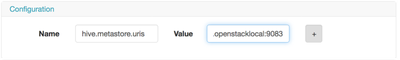
Now in WFM, you add that into the configuration section of the Pig action.
I also want to compress output coming from Mappers to improve performance for intermediate IO, I'm going to use property of Mapreduce called mapreduce.map.output.compress and set it to true
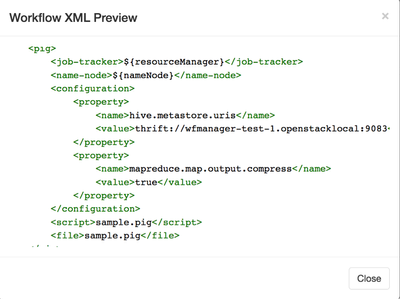
At this point, I'd like to see how I'm doing and I will preview the workflow in XML form. You can find it under workflow action.
This is also a good time to confirm your thrift URI and commonly forgotten property <script> and <file>.
Now finally, let's add the script to the wf directory. Use your favorite editor and paste the code for Pig and save file as sample.pig
set hcat.bin /usr/bin/hcat; sql show tables; A = LOAD 'data' USING org.apache.hive.hcatalog.pig.HCatLoader(); B = LIMIT A 1; DUMP B;
I have a Hive table called 'data' and that's what I'm going to load as part of Pig, I'm going to peek into the table and dump one relation to console. In the 2nd line of the script, I'm also executing a Hive "show tables;" command.
I also recommend to execute this script manually to make sure it works, command for it is
pig -x tez -f sample.pig –useHCatalog
Once it executes, you can see the output on the console, for brevity, I will only show the output we're looking for
2017-02-12 14:29:52,403 [main] INFO org.apache.pig.tools.grunt.GruntParser - Going to run hcat command: show tables; OK data wfd 2017-02-12 14:30:09,205 [main] INFO org.apache.pig.backend.hadoop.executionengine.util.MapRedUtil - Total input paths to process : 1 (abc,xyz)
Notice the output of show tables and then (abc,xyz) that's the data I have in my 'data' table.
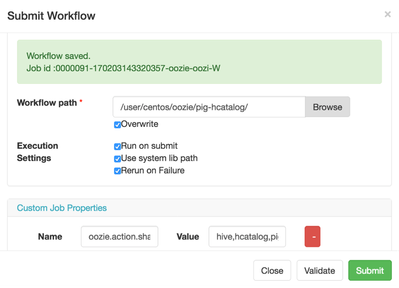
Finally, upload the file to wf directory. Save the wf first in WFM to create directory or point wf to an existing directory with the script in it.
hdfs dfs -put sample.pig oozie/pig-hcatalog/
We are finally ready to execute the wf. As the last step, we need to tell wf that we're going to use Pig with Hive and HCatalog and we need to add a property oozie.share.lib.for.pig=hive,pig,hcatalog. This property tells Oozie that we need to use more than just Pig libraries to execute the action.
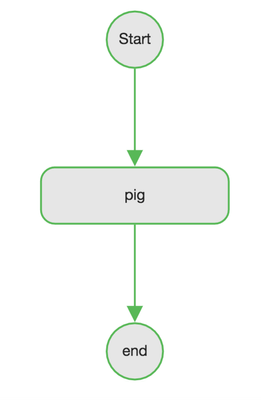
Let's check the status of the wf, click the Dashboard button. Luckily wf succeeded.
Let's click on the job and go to flow graph tab. All nodes appear in green, means it succeeded but we already knew that.
Navigate to action tab, we'll be able to drill to Resource Manager job history from that tab.
Let's click the arrow facing up to continue to RM. Go through the logs in job history and in stdout log you can find the output, we're looking for output of show tables and output of dump command.
Looks good to me. Thanks!
Created on 12-06-2017 04:56 PM
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Report Inappropriate Content
I'm trying to follow your tutorial, but I encountered problems:
- first: it didn't find Hive Metastore.class, so I added the .jar into pig directory
- now I'm obtaining in the stdout this:
2017-12-05 15:49:25,008 [ATS Logger 0] INFO org.apache.hadoop.yarn.client.api.impl.TimelineClientImpl - Exception caught by TimelineClientConnectionRetry, will try 1 more time(s). Message: java.net.ConnectException: Connessione rifiutata (Connection refused) 2017-12-05 15:49:26,011 [ATS Logger 0] INFO org.apache.pig.backend.hadoop.PigATSClient - Failed to submit plan to ATS: Failed to connect to timeline server. Connection retries limit exceeded. The posted timeline event may be missing Heart beat Heart beat<br>
My pig code is:
r = LOAD 'file.txt' using PigStorage(';') AS (
id:chararray,
privacy_comu:int,
privacy_util:int
);
r_clear = FOREACH r GENERATE TRIM($0) as id, $2 as privacy_trat,$3 as privacy_comu;
STORE r_clear INTO 'db.table' USING org.apache.hive.hcatalog.pig.HCatStorer();
I don't know what setup I need to work to pig with HCatalog and Oozie, could you suggest me any solutions about? @Artem Ervits
Thanks
Created on 01-25-2018 10:00 PM
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Report Inappropriate Content
Hi,
Thank you for providing these examples. I went through this one but could not make it work. the Job is being Killed for some reason. Looking at the LogError of the workflow, the following is the message I got:
USER[admin] GROUP[-] TOKEN[] APP[pigwf] JOB[0000006-180125094008677-oozie-oozi-W] ACTION[0000006-180125094008677-oozie-oozi-W@pig_1] Launcher ERROR, reason: Main class [org.apache.oozie.action.hadoop.PigMain], exit code [2]
Also, I looked at /var/log/oozie/oozie-error.log and I got the following message:
2018-01-25 11:20:07,800 WARN ParameterVerifier:523 - SERVER[my.server.com] USER[-] GROUP[-] TOKEN[-] APP[-] JOB[-] ACTION[-] The application does not define formal parameters in its XML definition 2018-01-25 11:20:07,808 WARN LiteWorkflowAppService:523 - SERVER[my.server.com] USER[-] GROUP[-] TOKEN[-] APP[-] JOB[-] ACTION[-] libpath [hdfs://my.server.com:8020/tmp/data/lib] does not exist 2018-01-25 11:20:30,527 WARN PigActionExecutor:523 - SERVER[PD-Hortonworks-DATANODE2.network.com] USER[admin] GROUP[-] TOKEN[] APP[pigwf] JOB[0000004-180125094008677-oozie-oozi-W] ACTION[0000004-180125094008677-oozie-oozi-W@pig_1] Launcher ERROR, reason: Main class [org.apache.oozie.action.hadoop.PigMain], exit code [2]
I ran the same script as you suggested and also tested it on shell which got me the result I was looking for. Also, I tested the oozie workflow with your part1 tutorial "making a shell command" and it worked. Also, I checked the workflow.xml files and everything looks like yours.
Could you please help me find what my problem is?
Thanks,
Sam