Community Articles
- Cloudera Community
- Support
- Community Articles
- Image Data Flow for Industrial Imaging
- Subscribe to RSS Feed
- Mark as New
- Mark as Read
- Bookmark
- Subscribe
- Printer Friendly Page
- Report Inappropriate Content
- Subscribe to RSS Feed
- Mark as New
- Mark as Read
- Bookmark
- Subscribe
- Printer Friendly Page
- Report Inappropriate Content
Created on 01-04-2019 03:24 AM - edited 08-17-2019 05:03 AM
Image Data Flow for Industrial Imaging
OBJECTIVE:
Ingest and store manufacturing quality assurance images, measurements, and metadata in a cost-effective and simple-to-retrieve-from platform that can provide analytic capability in the future.
OVERVIEW:
In high-speed manufacturing, imaging systems may be used to identify material imperfections, monitor thermal state, or identify when tolerances are exceeded. Many commercially-available systems automate measurement and reporting of specific tests, but combining results from multiple instrumentation vendors, longer-term storage, process analytics, and comprehensive auditability require different technology.
Using HDF’s NiFi and HDP’s HDFS, Hive or Hbase, and Zeppelin, one can build a cost-effective and performant solution to store and retrieve these images, as well as provide a platform for machine learning based on that data.
Sample files and code, including the Zeppelin notebook, can be found on this github repository: https://github.com/wcbdata/materials-imaging
PREREQUISITES:
HDF 3.0 or later (NiFi 1.2.0.3+)
HDP 2.6.5 or later (Hadoop 2.6.3+ and Hive 1.2.1+)
Spark 2.1.1.2.6.2.0-205 or later
Zeppelin 0.7.2+
STEPS:
- Get the files to a filesystem accessible to NiFi. In this case, we are assuming the source system can get the files to a local directory (e.g., via an NFS mount).
- Ingest the image and
data files to long-term storage
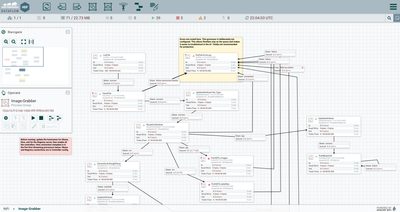
- Use a ListFile processor to scrape the directory. In this example, the collected data files are in a root directory, with each manufacturing run’s image files placed in a separate subdirectory. We’ll use that location to separate out the files later
- Use a FetchFile to pull the files listed in the flowfile generated by our ListFile. FetchFile can move all the files to an archive directory once they have been read.
- Since we’re using a
different path for the images versus the original source data, we’ll split the
flow using an UpdateAttribute to store the file type, then route them to two
PutHDFS processors to place them appropriately. Note that PutHDFS_images uses the
automatically-parsed original ${path} to reproduce the source folder structure.
- Parse the data files
to make them available for SQL queries
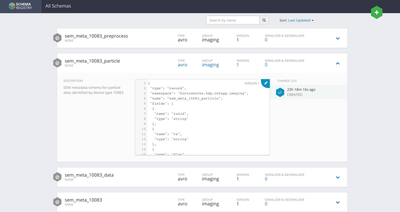
- Beginning with only the csv flowfiles, the ExtractGrok processor is used to pick one field from the second line of the flowfile (skipping the header row). This field is referenced by expression language that sets the schema name we will use to parse the flowfile.
- A RouteOnAttribute processor checks the schema name using a regex to determine whether it the flowfile format is one that requires additional processing to parse. In the example, flowfiles identified as the “sem_meta_10083” schema are routed to the processor group “Preprocess-SEM-particle.” This processor group contains the steps for parsing nested arrays within the csv flowfile.
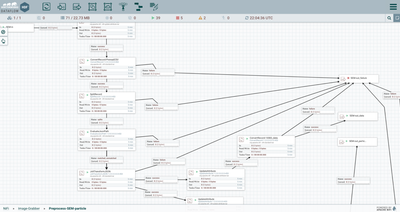
- Within the
“Preprocess-SEM-particle” processor group, the flowfile is parsed using a
temporary schema. A temporary schema can be helpful to parse some sections of a
flowfile row (or tuple) while leaving others for later processing.
- The flowfile is split into individual records by a SplitRecord processor. SplitRecord is similar to a SplitJSON or SplitText processor, but it uses NiFi’s record-oriented parsers to identify each record rather than relying strictly on length or linebreaks.
- A JoltTransform uses the powerful JOLT language to parse a section of the csv file with nested arrays. In this case, a semicolon-delimited array of comma-separated values is reformatted to valid JSON then split out into separate flowfiles by an EvaluateJSONPath processor. This JOLT transform uses an interesting combination of JOLT wildcards and repeated processing of the same path to handle multiple possible formats.
- Once formatted by a
record-oriented processor such as ConvertRecord or SplitRecord, the flowfile
can be reformatted easily as Avro, then inserted into a Hive table using a
PutHiveStreaming processor. PutHiveStreaming can be configured to ignore extra
fields in the source flowfile or target Hive table so that many overlapping
formats can be written to a table with a superset of columns in Hive. In this
example, the 10083-formatted flowfiles are inserted row-by-row, and the
particle and 10021-formatted flowfiles are inserted in bulk.
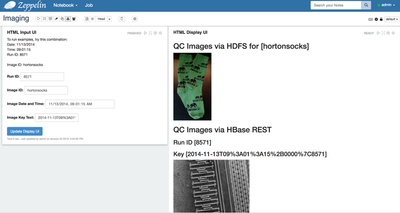
- Create a simple interface to retrieve individual images for review. The browser-based Zeppelin notebook can natively render images stored in SQL tables or in HDFS.
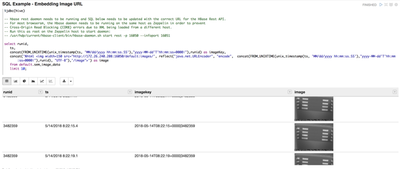
- The notebook begins with some basic queries to view the data loaded from the imaging subsystems.
- The first example
paragraph uses SQL to pull a specific record from the manufacturing run, then
looks for the matching file on HDFS by its timestamp.
- The second set of
example paragraphs use an HTML/Angular form to collect the information, then
display the matching image.
- The third set of
sample paragraphs demonstrates how to obtain the image via Scala for analysis
or display.
RELATED POSTS:
FUTURE POSTS:
- Storing microscopy data in HDF5/USID Schema to make it available for analysis using standard libraries
- Applying TensorFlow to microscopy data for image analysis