Community Articles
- Cloudera Community
- Support
- Community Articles
- Visualize patients' complaints to their doctors us...
- Subscribe to RSS Feed
- Mark as New
- Mark as Read
- Bookmark
- Subscribe
- Printer Friendly Page
- Report Inappropriate Content
- Subscribe to RSS Feed
- Mark as New
- Mark as Read
- Bookmark
- Subscribe
- Printer Friendly Page
- Report Inappropriate Content
Created on
03-01-2016
12:22 AM
- edited on
02-20-2020
12:39 AM
by
SumitraMenon
Summary
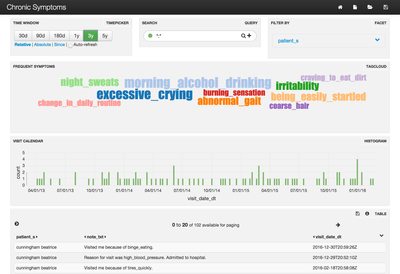
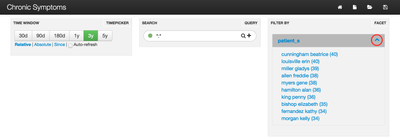
Because patients visit many doctors, trends in their ailments and complaints may be difficult to identify. The steps in this article will help you address exactly this problem by creating a TagCloud of the most frequent complaints per patient. Below is a sample:
We will generate random HL7 MDM^T02 (v2.3) messages that contain a doctor's note about a fake patient and that patient's fake complaint to their doctor. Apache NiFi will be used to parse these messages and send them to Apache Solr. Finally Banana is used to create the visual dashboard.
In the middle of the dashboard is a TagCloud where the more frequently mentioned symptoms for a selected patient appear larger than others. Because this project relies on randomly generated data, some interesting results are possible. In this case, I got lucky and all the symptoms seem related to the patient's most frequent complaint: Morning Alcohol Drinking. The list of all possible symptoms comes from Google searches.
Summary of steps
- Download and install the HDP Sandbox
- Download and install the latest NiFi release
- Download the HL7 message generator
- Create a Solr dashboard to visualize the results
- Create and execute a new NiFi flow
Detailed Step-by-step guide
1. Download and install the HDP Sandbox
Download the latest (2.3 as of this writing) HDP Sandbox here. Import it into VMware or VirtualBox, start the instance, and update the DNS entry on your host machine to point to the new instance’s IP. On Mac, edit /etc/hosts, on Windows, edit %systemroot%\system32\drivers\etc\ as administrator and add a line similar to the below:
192.168.56.102 sandbox sandbox.hortonworks.com
2. Download and install the latest NiFi release
Follow the directions here. These were the steps that I executed for 0.5.1
wget http://apache.cs.utah.edu/nifi/0.5.1/nifi-0.5.1-bin.zip -O /tmp/nifi-0.5.1-bin.zip cd /opt/ unzip /tmp/nifi-0.5.1-bin.zip useradd nifi chown -R nifi:nifi /opt/nifi-0.5.1/ perl -pe 's/run.as=.*/run.as=nifi/' -i /opt/nifi-0.5.1/conf/bootstrap.conf perl -pe 's/nifi.web.http.port=8080/nifi.web.http.port=9090/' -i /opt/nifi-0.5.1/conf/nifi.properties /opt/nifi-0.5.1/bin/nifi.sh start
3. Download the HL7 message generator
A big thank you to HAPI for their excellent library to parse and create HL7 messages on which my code relies. The generator creates a very simple MDM^T02 that includes an in-line note from a doctor. MDM stands for Medical Document Management, and T02 specifies that this is a message for a new document. For more details about this message type read this document. Here is a sample message for Beatrice Cunningham:
MSH|^~\&|||||20160229002413.415-0500||MDM^T02|7|P|2.3 EVN|T02|201602290024 PID|1||599992601||cunningham^beatrice^||19290611|F PV1|1|O|Burn center^60^71 TXA|1|CN|TX|20150211002413||||||||DOC-ID-10001|||||AU||AV OBX|1|TX|1001^Reason For Visit: |1|Evaluated patient for skin_scaling. ||||||F
As a pre-requisite to executing the code, we need to install Java 8. Execute this on the Sandbox:
yum -y install java-1.8.0-openjdk.x86_64
Now, download the pre-build jar file that has the HL7 generator and execute it to create a single message in /tmp/hl7-messages. I chose to store the jar file in /var/ftp/pub because my IDE uploads files during code development. If you change this directory, also change it in the NiFi flow.
mkdir -p /var/ftp/pub cd /var/ftp/pub wget https://raw.githubusercontent.com/vzlatkin/DoctorsNotes/master/target/hl7-generator-1.0-SNAPSHOT-sha... mkdir -p /tmp/hl7-messages/ /usr/lib/jvm/jre-1.8.0/bin/java -cp hl7-generator-1.0-SNAPSHOT-shaded.jar com.hortonworks.example.Main 1 /tmp/hl7-messages chown -R nifi:nifi /tmp/hl7-messages/
4. Create a Solr dashboard to visualize the results
Now we need to configure Solr to ignore some words that don't add value. We do this by modifying stopwords.txt
cat <<EOF > /opt/hostname-hdpsearch/solr/server/solr/configsets/data_driven_schema_configs/conf/stopwords.txt adjustments Admitted because blood changes complained Discharged Discussed Drew Evaluated for hospital me medication of patient Performed Prescribed Reason Recommended Started tests The to treatment visit Visited was EOF
Next, we download the custom dashboard and start Solr in cloud mode
export JAVA_HOME=/usr/lib/jvm/java-1.7.0-openjdk.x86_64 wget "https://raw.githubusercontent.com/vzlatkin/DoctorsNotes/master/other/Chronic%20Symptoms%20(Solr).json" -O /opt/hostname-hdpsearch/solr/server/solr-webapp/webapp/banana/app/dashboards/default.json /opt/hostname-hdpsearch/solr/bin/solr start -c -z localhost:2181 /opt/hostname-hdpsearch/solr/bin/solr create -c hl7_messages -d data_driven_schema_configs -s 1 -rf 1
5. Create and execute a new NiFi flow
- Start by downloading this NiFi template to your host machine.
- To import the template, open the NiFi UI
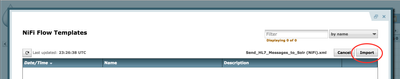
- Next, open Templates manager:
-
Click "Browse", then find the template on your local machine, click "Import", and close the Template Window.
-
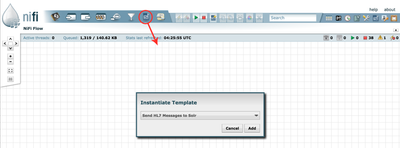
Drag and drop to instantiate a new template:
-
Double click the new process group called HL7, and start all of the processes. To do so, hold down the Shift-key, and select all of the processes on the screen. Then click the "Start" button:
Here is a quick walk through of the processes starting in the top-left corner. First, we use ListFile process to get a directory listing from /tmp/hl7-messages. Second, the FetchFile process reads each file one-by-one, passes the contents to the next step, and deletes if successful. Third, the text file is parsed as an HL7 formatted message. Next, the UpdateAttribute and AttributesToJSON processes get the contents ready for insertion into Solr. Finally, we use the PutSolrContentStream process to add new documents via Solr REST API. The remaining two processes on the very bottom are for spawning the custom Java code and logging details for troubleshooting.
Conclusion
Now open the Banana UI. You should see a dashboard that looks similar to the screenshot in the beginning of this article. You can see how many messages have been processed by clicking the link in the top-right panel called "Filter By".
Troubleshooting
If you are not seeing any data in Solr/Banana, then reload the page. Also perform a search via this page to validate that results are being indexed via Solr correctly.
Full source code is located in GitHub.
Created on 03-01-2016 12:25 AM
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Report Inappropriate Content
Bravo Vladimir! Do you have a github repo? I'd like to contribute.
Created on 03-01-2016 12:52 AM
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Report Inappropriate Content
Yes, I should have added a link to GitHub: https://github.com/vzlatkin/DoctorsNotes
Created on 03-01-2016 05:41 PM
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Report Inappropriate Content
@Vladimir Zlatkin I just tried out your demo, there are a few things I got stuck on. I was not able to download the custom dashboard via your URL due to the parenthesis. I wrapped the URL in double quotes and it worked. Then I was having issues creating a collection hl7_messages, is creating solr directory in zookeeper necessary? I created it anyway but I also had to refer to Ali's nifi twitter tutorial for reference in terms of Solr. here's link https://community.hortonworks.com/content/kbentry/1282/sample-hdfnifi-flow-to-push-tweets-into-solrb... I finally was able to create hl7_messages collection but upon template execution, I'm getting errors in Solr that the collection doesn't exist, hence asking you whether zk step is necessary? Will do further digging.
- Ensure no log files owned by root (current sandbox version has files owned by root in log dir which causes problems when starting solr)
- chown -R solr:solr /opt/lucidworks-hdpsearch/solr
- Run solr setup steps as solr user
- su solr
- Setup the Banana dashboard by copying default.json to dashboard dir
- cd /opt/lucidworks-hdpsearch/solr/server/solr-webapp/webapp/banana/app/dashboards/
- mv default.json default.json.ori
Created on 03-01-2016 05:46 PM
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Report Inappropriate Content
@Artem Ervits Thanks for giving this tutorial a try. If you are getting the errors on an HDP Sandbox, would you send me the .vmdk file? I'll take a look and see what needs to change in the tutorial.
Created on 03-01-2016 05:49 PM
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Report Inappropriate Content
I wouldn't know where to drop a large vmdk like that? @Vladimir Zlatkin I'll give it a go once more before going the PS support route ;).
Created on 03-02-2016 03:59 AM
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Report Inappropriate Content
Problems fixed. There is no longer a step to chroot Solr directory in Zookeeper.
Created on 06-09-2016 01:19 PM
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Report Inappropriate Content
Great Demo Valdimir, much appreciated. I am just digging into the banana facet code a little, it looks the filter by query has one patient "cunningham" in the query string. If I figure it out I will post, lol.
Ahh, you just need to X out cunningham and then the list appears, got it, after a little solr learning
Created on 12-06-2017 04:07 PM
- Mark as Read
- Mark as New
- Bookmark
- Permalink
- Report Inappropriate Content
https://community.hortonworks.com/articles/149910/handling-hl7-records-part-1-hl7-ingest.html
Attribute Name Cleaner (Needed for messy C-CDA and HL7 attribute names)