Support Questions
- Cloudera Community
- Support
- Support Questions
- Best way to select distinct values from multiple c...
- Subscribe to RSS Feed
- Mark Question as New
- Mark Question as Read
- Float this Question for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Subscribe to RSS Feed
- Mark Question as New
- Mark Question as Read
- Float this Question for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Best way to select distinct values from multiple columns using Spark RDD?
- Labels:
-
Apache Spark
Created 12-10-2015 01:37 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
I'm trying to convert each distinct value in each column of my RDD, but the code below is very slow. Is there any alternative?
Data is both numeric and categorical (string).
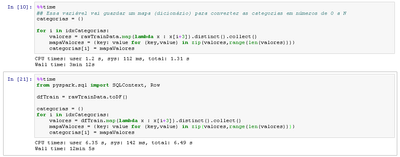
categories = {}
for i in idxCategories: ##idxCategories contains indexes of rows that contains categorical data
distinctValues = rawTrainData.map(lambda x : x[i]).distinct().collect()
valuesMap = {key: value for (key,value) in zip(distinctValues, range(len(valores)))}
categories[i] = valuesMap
Created 12-10-2015 02:28 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
Data Frames are supposed to be faster than Python RDD operations, check slide 20 of this presentation:
Could you try code below and check if it's faster?
from pyspark.sql import SQLContext, Row
input_file = "hdfs:///tmp/your_text_file"
raw_rdd = sc.textFile(input_file)
csv_rdd = raw_rdd.map(lambda x: x.split(","))
row_data = csv_rdd.map(lambda p: Row(
field1=p[0],
field2=p[1],
field3=p[2]
)
)
df = sqlContext.createDataFrame(row_data)
categories = {}
idxCategories = [0,1,2]
for i in idxCategories: ##idxCategories contains indexes of rows that contains categorical data
distinctValues = df.map(lambda x : x[i]).distinct().collect()
categories[i] = distinctValues
print categories[0]
print categories[1]
print categories[2]
Created 12-10-2015 02:23 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
A few clarifying questions about rawTrainData:
- How is this RDD generated?
- Is it cached?
- how many partitions does it have?
Also, what is the variable "valores"?
Created 12-10-2015 03:22 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
- RDD is read from CSV and split into list
- rawTrainData is cached
- It have 2 partitions at same node. The file is not large. 220 MB.
- I edited original code to translate to English. Valores = distincValues
Created 12-10-2015 02:28 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
Data Frames are supposed to be faster than Python RDD operations, check slide 20 of this presentation:
Could you try code below and check if it's faster?
from pyspark.sql import SQLContext, Row
input_file = "hdfs:///tmp/your_text_file"
raw_rdd = sc.textFile(input_file)
csv_rdd = raw_rdd.map(lambda x: x.split(","))
row_data = csv_rdd.map(lambda p: Row(
field1=p[0],
field2=p[1],
field3=p[2]
)
)
df = sqlContext.createDataFrame(row_data)
categories = {}
idxCategories = [0,1,2]
for i in idxCategories: ##idxCategories contains indexes of rows that contains categorical data
distinctValues = df.map(lambda x : x[i]).distinct().collect()
categories[i] = distinctValues
print categories[0]
print categories[1]
print categories[2]
Created on 12-10-2015 03:18 PM - edited 08-19-2019 05:39 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
4x slower 😞 I used .toDF() instead of your code. Is there any difference?
Created 12-10-2015 05:16 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
You could load your csv directly, but I tested here and indeed distinct is take much longer with data frames.
Can you describe your environment?
- hortonworks version
- spark version
- hardware configuration
- spark mode (localmode or spark on yarn)
Lastly, if you have enough cores/processor and as your file is small, spark might be choosing a low level of parallelism. you can try it increasing parallelism, like this:
distinctValues = rawTrainData.map(lambda x : x[i]).distinct(numPartitions = 15).collect()
me fala se ficou mais rápido 🙂
Created 12-10-2015 08:28 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
before calling this routine, I introduced the code bellow and exec time reduced to 1m8s. 3x improvement.
rawTrainData = rawTrainData.repartition(8) rawTrainData.cache()
But introducing numPartitions=15 inside distinct method does not affect the result.
I'm running Spark 1.3.1 into standalone mode (spark://host:7077) with 12 cores and 20 GB per node allocated to Spark. The hardware is virtual, but I know it`s a top hardware. The cluster has 4 nodes (3 spark workers)
Created 12-11-2015 12:48 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
Awesome! You can check current number of partitions with command below:
print csv_rdd.getNumPartitions()
Created 02-02-2016 01:52 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
@Vitor Batista can you accept the best answer to close this thread or post your own solution?