Hi!
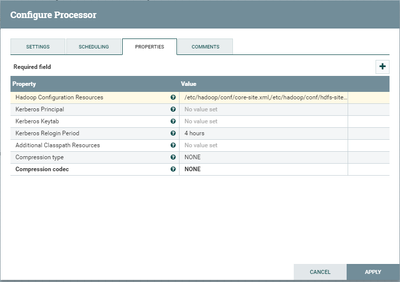
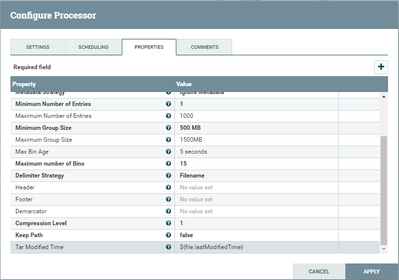
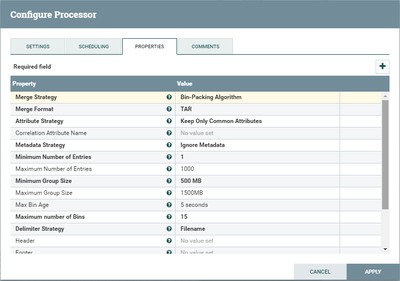
I have a dataflow in which I create a sequence file from multiple files and load it to hdfs.
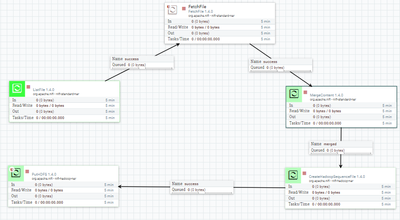
Unfortunately I cannot correctly read the generated file in Spark.
For example, I generate 5 txt files:
1.txt
1
2.txt
2
22
3.txt
3
33
333
4.txt
4
44
444
4444
5.txt
5
55
555
5555
55555
and create from those files the new sequence file.
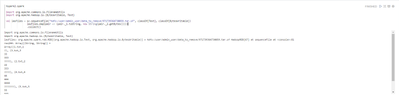
After that I try to read the resulting file:
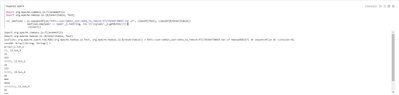
We can see there are corrupted or trash characters in output (they are zero bytes).
How I can get rid from those unnecessary bytes?
Some additional screenshots are attached.