Support Questions
- Cloudera Community
- Support
- Support Questions
- Re: Unable to start processor NIFI,
- Subscribe to RSS Feed
- Mark Question as New
- Mark Question as Read
- Float this Question for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Subscribe to RSS Feed
- Mark Question as New
- Mark Question as Read
- Float this Question for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Unable to start processor NIFI,
- Labels:
-
Apache NiFi
Created on 04-26-2016 08:32 PM - edited 08-19-2019 03:49 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
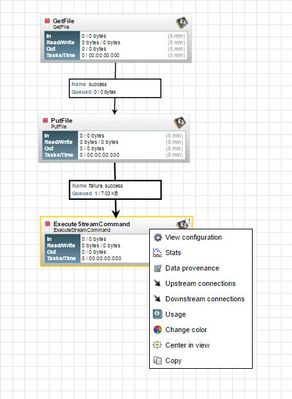
Unable start processor after once stopping it, upon right click no start or edit configuration .
Files gets queued up Execute stream processor wont pick them up.
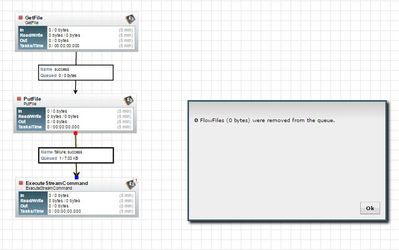
Unable to empty the queue using empty queue. Requires nifi to restarted.
Hardware configuration :
8 cores
60 GB Ram
160gb Harddisk
Created 04-26-2016 08:40 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
Hello
Unfortunately this means the process NiFi was told to execute has not returned. In such a case there is an outstanding thread and we intentionally prevent additional instances from being started until this one is dealt with. From the stacktrace you provided we see
"Timer-Driven Process Thread-9" Id=69 RUNNABLE (in native code) at java.io.FileInputStream.readBytes(Native Method) at java.io.FileInputStream.read(FileInputStream.java:272) at java.io.BufferedInputStream.read1(BufferedInputStream.java:273) at java.io.BufferedInputStream.read(BufferedInputStream.java:334) - waiting on java.lang.UNIXProcess$ProcessPipeInputStream@e6f0a2f
This tells us we're sitting and waiting for that command to do something (finish, respond with data). It appears to be in a hung state. You'll need to restart NiFi and try to assess why that command isn't doing anything. What command are you trying to run? It might be that for your case the 'ExecuteProcess' processor is a better fit.
Thanks
Joe
Created 04-26-2016 08:40 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
Hello
Unfortunately this means the process NiFi was told to execute has not returned. In such a case there is an outstanding thread and we intentionally prevent additional instances from being started until this one is dealt with. From the stacktrace you provided we see
"Timer-Driven Process Thread-9" Id=69 RUNNABLE (in native code) at java.io.FileInputStream.readBytes(Native Method) at java.io.FileInputStream.read(FileInputStream.java:272) at java.io.BufferedInputStream.read1(BufferedInputStream.java:273) at java.io.BufferedInputStream.read(BufferedInputStream.java:334) - waiting on java.lang.UNIXProcess$ProcessPipeInputStream@e6f0a2f
This tells us we're sitting and waiting for that command to do something (finish, respond with data). It appears to be in a hung state. You'll need to restart NiFi and try to assess why that command isn't doing anything. What command are you trying to run? It might be that for your case the 'ExecuteProcess' processor is a better fit.
Thanks
Joe
Created 04-26-2016 09:07 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
#!/usr/local/bin/python2.7
import os,gzip,sys,csv,shutil,time,subprocess
#Read the TagListDataOnly.csv file
ifile = open(sys.argv[3],'rU')
reader = csv.reader(ifile, delimiter=',')
#Building dictionary with key as code and value as metric name
mydict = dict((str(rows[1]),str(rows[3])) for rows in reader)
pattern = '%d-%m-%Y %H:%M:%S.%f'
finallines=[]
inputfilename=str(sys.argv[6])+str(sys.argv[2])
serialno=sys.argv[2].split('_')[-2][:-1]+'9'
with gzip.open(inputfilename,'r') as data:
for line in data:
elements = line.split(',')
epoch = int(time.mktime(time.strptime(elements[3].rstrip(), pattern)))
origMetricName = mydict[elements[0]]
if int(elements[2]) == 192:
quality='0'
else:
quality='1'
if elements[1].rstrip().lstrip('-').replace('.','',1).isdigit():
if origMetricName.rsplit('_',-1)[-1].isdigit():
metricname ='_'.join(mydict[elements[0]].rsplit('_',-1)[0:-1])
finalline = metricname+' '+str(epoch)+' '+elements[1].rstrip()+' SerialNo='+sys.argv[2].split('_')[-2][:-1]+'9'+' Index='+origMetricName.split('_')[-1]+' DataQuality='+quality+ ' DataType=RAW DataSource=OPC \n'
else:
metricname = origMetricName
finalline = metricname+' '+str(epoch)+' '+elements[1].rstrip()+' SerialNo='+sys.argv[2].split('_')[-2][:-1]+'9'+' DataQuality='+quality+' DataType=RAW DataSource=OPC \n'
finallines.append(finalline)
else:
pass
outputcsv=str(sys.argv[4])+str(sys.argv[2]).split('.')[0]+'9'+"_Changed.csv"
outputcsvgzdir=str(sys.argv[5])+'/'+serialno+'/Sensor/'
if not os.path.exists(outputcsvgzdir):
os.makedirs(outputcsvgzdir)
outputcsvgz=str(sys.argv[5])+'/'+serialno+'/Sensor/'+str(sys.argv[2]).split('.')[0]+'9'+"_Changed.csv.gz"
target = open(outputcsv,"w")
target.writelines(finallines)
target.close()
with open(outputcsv, 'rb') as f_in, gzip.open(outputcsvgz, 'wb') as f_out:
shutil.copyfileobj(f_in, f_out)
os.remove(outputcsv)
#os.remove(inputfilename)
command="/opt/opentsdb/build/tsdb import --auto-metric --skip-errors --zkquorum=172.31.19.88:2181 --zkbasedir=/hbase-unsecure %s" % outputcsvgz
process = subprocess.Popen(command,shell=True, stdout=subprocess.PIPE, stderr=subprocess.PIPE);
process.wait();
output, err = process.communicate()
debug=open('/import/debug/debug-'+sys.argv[2].split('.')[0]+'9'+'.txt','w')
debug.write('---err txt---\n')
debug.write(err)
debug.write('---op txt---\n')
debug.write(output)
debug.write(outputcsvgz)
debug.close()
print process.returncode;
HI Joe,
Thank you on quick response.
I am trying to perform ETL process which tranforms data and bulk ingest data into opentsdb. below are python script and NIFI xml file etl-1.xml
Thanks
Raghu
Created 04-27-2016 04:34 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
Does the python script successfully finish when manually run outside of Nifi?
Created 06-08-2018 08:13 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
Hi Raghu
Can you please help me in understanding how to load the data into OpenTSDB using Nifi alone?