Support Questions
- Cloudera Community
- Support
- Support Questions
- Re: Decompress the GZ file
- Subscribe to RSS Feed
- Mark Question as New
- Mark Question as Read
- Float this Question for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
- Subscribe to RSS Feed
- Mark Question as New
- Mark Question as Read
- Float this Question for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Decompress the GZ file
- Labels:
-
Apache Hadoop
-
Apache NiFi
Created on 04-03-2019 11:48 AM - edited 08-17-2019 04:11 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
Hi All,
i'm trying to decompress a gz file from HDFS location and put it in another location. But the FetchHDFS is not fetching the file at all?
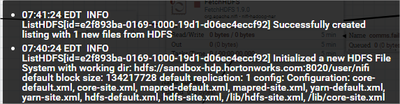
This is the ListHDFS warning I'm getting:
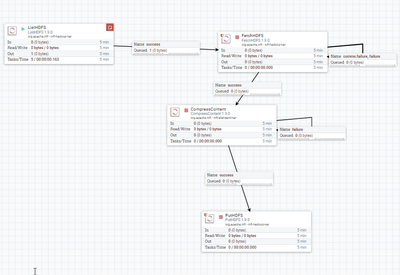
My Flow:
And my configuration:
ListHDFS https://snag.gy/gWoLej.jpg
FetchHDFS https://snag.gy/N1IEfB.jpg
As I understan, the listing part works: Successfuly created listing with 1 new files from HDFS. But the FetchHDFS is not doing anything. Did I miss something in the configuration?
Thanks!
N
Created on 04-03-2019 02:17 PM - edited 08-17-2019 04:11 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
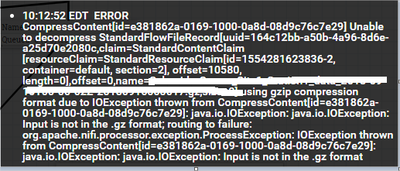
I managed to get the file to the decompress processor, but got an error saying the file is not in gz format. and it is.
Created 04-03-2019 11:26 PM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
Make sure your fetched file have .gz extention in filename, if yes then check is the .gz file is in valid, by uncompress in the shell using gunzip command.
Created 04-05-2019 01:57 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
I tried on my local instance and everything works as expected.
-
If you have .gz file in local FS then try to fetch the file ListFile+FetchFile from your local FS(instead of HDFS) and check are you able to fetch the whole file without any issues?.
-
Move Local file to HDFS using the below command.
hadoop fs -put <local_File_path> <hdfs_path>
then check are u able to get the file size as 371kb in hdfs?
-
If yes then try to run
ListHDFS+FetchHDFS processors to fetch the newly moved file into HDFS directory.
-
Some threads related to similar issue.
https://community.hortonworks.com/questions/106925/error-when-sending-data-to-api-from-nifi.html
Created 04-04-2019 07:30 AM
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Report Inappropriate Content
@Shu I was able to decompress it in shell. it's not corrupt.
I've also noticed, that when I moved (list, movehdfs) the file with NiFi, the file was moved but with 0 kb. but when I copied it in shell with my user it had 371 kB. I was not able to fetch the 371 kb file because of the https://snag.gy/jExmbl.jpg and https://snag.gy/eVEjG7.jpg error.
Any ideas?